More Information
Submitted: December 06, 2023 | Approved: December 15, 2023 | Published: December 18, 2023
How to cite this article: Dasaradharami Reddy K, Anusha S, Chandrakala P. A Mini Review of Newly Identified Omicron Sublineages. Arch Case Rep. 2023; 7: 066-076.
DOI: 10.29328/journal.acr.1001082
Copyright License: © 2023 Dasaradharami Reddy K, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: COVID-19; Concerning variants; Omicron sublineages
A Mini Review of Newly Identified Omicron Sublineages
Dasaradharami Reddy K1*, Anusha S2 and Palem Chandrakala2
1School of Computer Science Engineering & Information Systems, Vellore Institute of
Technology, India
2Assistant Professor, Department of Computer Science and Engineering, NBKR
Institute of Science and Technology, Vidyanagar, India
*Address for Correspondence: Dasaradharami Reddy K, School of Computer Science Engineering & Information Systems, Vellore Institute of Technology, India, Email: [email protected]
The ongoing COVID-19 pandemic has seen the evolution of the SARS-CoV-2 virus, resulting in the emergence of various concerning variants with unique biological characteristics. As the pandemic continues, it will be crucial to promptly evaluate the potential of any new variant to cause severe illness. The severity of the latest Omicron sublineages, including BA.5, XBB, BQ.1.18, BA.2, BA.2.75, and EG.5.1, is currently under assessment. This system provides valuable and essential information for rapidly assessing the threat posed by new versions of the virus.
As the COVID-19 pandemic has gone on for the past three years, the virus that causes the disease, SARS-CoV-2, has changed and created different versions. Some of these versions were called “variants of concern” by the World Health Organization (WHO) [1,2]. So far, the WHO has identified five variants of concern: Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and the new Omicron variant, also known as B.1.1.529, spreads much faster than the other four variants [3,4]. The threat of a new variant depends on how easily it spreads, how sick it makes people compared to other versions, and how well it can get around our immune defenses. These variants have changed because of many different mutations, especially in the spike protein of the virus, which is important for how it infects us. Some important mutations that make the variants more infectious are found in different versions like Alpha, Beta, Gamma, Delta, and Omicron [5-8]. Omicron seems to cause milder symptoms than other strains, but it spreads more easily and might not be stopped as well by vaccines, even though it doesn’t seem to be as deadly [9-11].
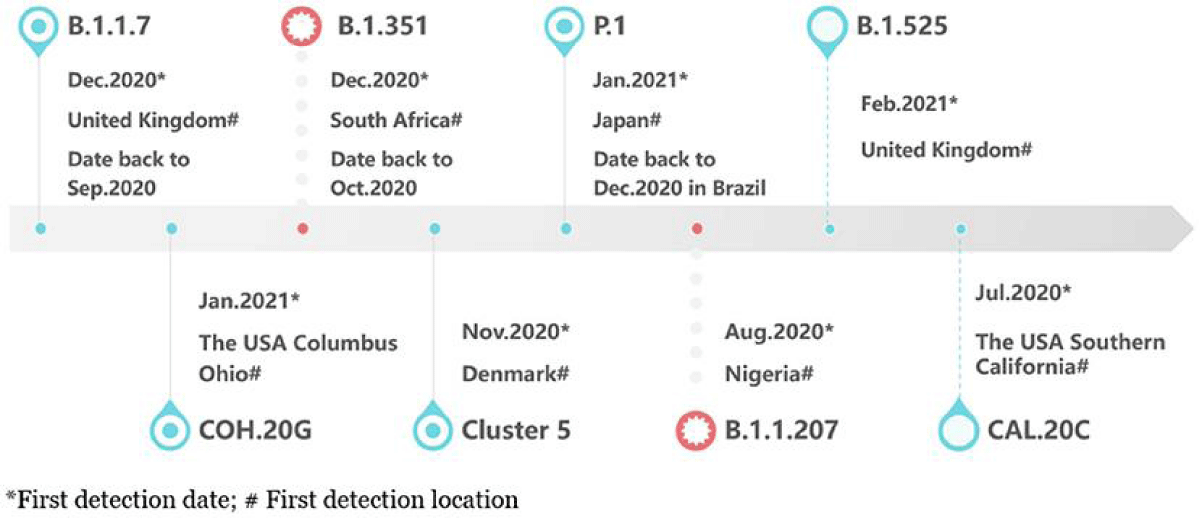
The Omicron variant of COVID-19 was first seen in South Africa and Botswana in November 2021 [12] (Figure 1). Since then, there have been over 130 million cases and 500,000 deaths worldwide. The Omicron variant has caused a big increase in COVID-19 cases, creating a new wave called the Omicron wave [13]. This wave has been much bigger than waves caused by other variants like Alpha and Beta (Figure 2). The Omicron variant has changed over time, creating different sub-types called sub-variants. These include BA.1, BA.2, BA.3, BA.4, BA.5, and a mix of BA.1 and BA.2 [14]. BA.1 was the most common globally, but BA.2 has become more common in many countries. BA.3 is not spreading much [15] (Figure 1). Two new sub-variants, BA.4 and BA.5, were found in South Africa in early 2022 and became the main types during the 5th wave of the pandemic there [16] (Figure 1). Omicron has more mutations than other variants, which helps it attach to human cells more strongly. It can also avoid some of the antibodies produced by vaccines or by people who have other variants of the virus [17-19].
Figure 1: The timeline explains how the Omicron variant of SARS-CoV-2 started and when its different sub-types emerged [59].
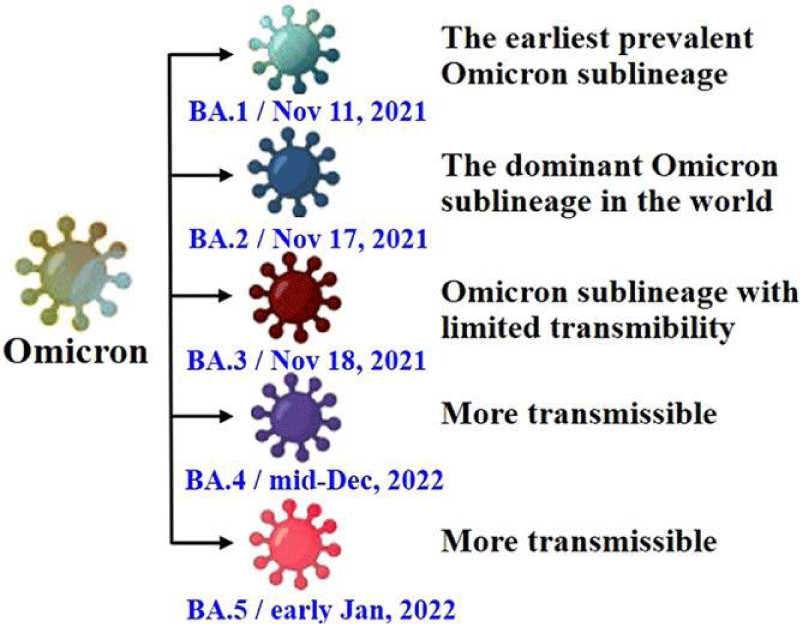
Figure 2: The diagram shows the characteristics of various sub-types of the Omicron variant, including BA.1, BA.2, BA.3, BA.4, and BA.5 [60].
The Omicron variant has many mutations in its genetic code. According to a report from the WHO in April 2022, there are five sub-types of the Omicron variant: BA.1, BA.2, BA.3, BA.4, and BA.5. These mutations have been found all over the world in different amounts (Figure 2) [20].
Kumar and colleagues used computer tools to study how infectious and harmful the S-glycoprotein of the BA.1 sub-type and its related sub-types (BA.1.1, BA.2, and BA.3) are. BA.1 has 39 genetic changes, while BA.1.1 has 40 changes [21]. BA.2 and BA.3 have 31 and 34 changes, respectively. There are 21 mutations that are common to all the evolved sub-types of the Omicron variant. Additionally, 11 usual changes have been found in the RBD of the Omicron variant and its evolved sub-types. The mutations in the spike protein of the BA.1 and BA.2 variants are only seen in the recently emerged BA.3 sublineage [22]. Data from https://outbreak.info/, accessed on December 14, 2023, shows that the BA.1 sublineage makes up 5% of cases in 161 countries, followed by the BA.1.1 variant at 17% in the same countries. The BA.2 sub-variant has seen a significant increase, accounting for 9% of reported cases across 163 different countries, while the prevalence of BA.3 cases was very low until May 2022 [21]. These variants have been found to evade the immune system and reduce the effectiveness of vaccines. The BA.1 sub-variant is more contagious than the previously dominant Delta variant, but infected individuals rarely need hospital care. The BA.2 sub-variant, due to the presence of the H78Y mutation, is more severe than the BA.1 sub-variant [23]. The latest transmission rates are attributed to the BA.3 sublineage, which lacks six mutations in its genome: L981F, G496S, ins214EPE, N856K, T547K, and S371L [22].
In early 2022, scientists discovered two new versions of the Omicron variant in South Africa, called BA.4 and BA.5. These new versions spread to many places around the world. By the end of 2021, the BA.1 variant became the main cause of the fourth wave, replacing the Delta variant. Then, by April 2022, the BA.1 variant was replaced by BA.2, which caused the fifth wave of COVID-19 [23,24]. These new variants are taking over from the older versions of Omicron. The spike proteins of these new versions are somewhat similar. BA.4 and BA.5 have extra mutations in the virus’s genetic material, similar to the B.1.429 variant, which was also seen in BA.2 [25]. These new versions can evade the body’s immune system. However, we don’t know much about how often people with BA.4 or BA.5 need to go to the hospital. One of the new versions, BA.2.12.1, can stop the antibodies from vaccines or previous infections with Omicron [26]. In short, BA.4, BA.5, and BA.2.12.1 are stronger and can avoid the body’s immune response [27].
Several new combinations of sub-variants are spreading in the community, like XD, XE, and XF. The XE variant, made up of BA.1 and BA.2, is especially concerning because it’s much more infectious than BA.2 [28-31]. XD is a mix of BA.1 and Delta, and XF is a mix of BA.1 and Delta from the UK. The XE variant has been called “stealth Omicron” by the WHO and is causing worry due to its high infectivity [31]. It has three new mutations not seen in BA.1 and BA.2. XD, first found in France, has a new mutation in the nsp2 gene, and XF has a unique change at the end of the nsp3 gene. Scientists are concerned about how severe infections caused by these new combinations might be [30].
Scientists have observed that many new variations of the virus have appeared after Omicron, like XBB, XBD, and XBF. We still need to study how these new variations work in the body [32].
SARS-CoV-2, the virus responsible for COVID-19, has undergone several mutations, leading to the emergence of different variants. These variants have raised concerns due to their potential impact on transmissibility, severity of illness, and effectiveness of vaccines and treatments. The Omicron variant, which was first identified in November 2021, has since given rise to multiple sub-lineages. These sublineages, designated as BA.1, BA.1.1, BA.2, and so on, represent distinct genetic variations within the Omicron variant. Each sublineage may possess unique mutations that could potentially affect the behaviour of the virus, including its transmissibility and ability to evade immunity.
The emergence of different variants of SARS-CoV-2, including the Omicron variant and its sublineages, has indeed raised significant concerns within the global health community. These variants have the potential to impact the transmissibility of the virus, the severity of illness it causes, and the effectiveness of existing vaccines and treatments. The Omicron variant, first identified in November 2021, has garnered attention due to its numerous mutations, which set it apart from earlier variants such as Delta. These mutations have the potential to alter the behaviour of the virus in several ways.
One of the primary concerns surrounding the Omicron variant and its sublineages is their potential impact on transmissibility. Certain mutations within these variants may enhance the virus’s ability to spread from person to person, leading to an increased rate of transmission within communities. This heightened transmissibility could potentially result in more rapid and widespread outbreaks of COVID-19. Additionally, the mutations present in the Omicron sublineages may also affect the severity of illness caused by the virus. While initial reports suggested that infections with the Omicron variant might be associated with milder symptoms compared to earlier variants, further research is needed to fully understand the impact of these mutations on disease severity.
Furthermore, the genetic variations within the Omicron sublineages have raised concerns about their potential to evade immunity, including immunity conferred by prior infection or vaccination. Certain mutations may enable the virus to partially evade the immune response, potentially reducing the effectiveness of existing vaccines and treatments. This has prompted ongoing research and surveillance to assess the ability of current vaccines to provide protection against the Omicron variant and its sublineages. It’s important to note that the situation surrounding SARS-CoV-2 variants, including Omicron and its sublineages, continues to evolve as new data and research become available. Continued genomic surveillance, research, and public health measures are essential for monitoring the spread and impact of these variants and for informing public health responses.
In summary, the emergence of the Omicron variant and its sublineages underscores the ongoing challenges posed by SARS-CoV-2 and the importance of global collaboration in monitoring, understanding, and responding to the evolution of the virus.
The Omicron variant of the SARS-CoV-2 virus has garnered significant attention due to its numerous mutations, particularly within the spike protein. The spike protein is crucial because it is the primary target of many COVID-19 vaccines. The presence of a large number of mutations in this region has raised concerns about the potential impact on vaccine effectiveness and the virus’s ability to evade immunity acquired through previous infection or vaccination. Understanding the implications of these sublineages is crucial for several reasons. Firstly, it is essential to inform public health measures. By closely monitoring and studying these sublineages, researchers and public health authorities can better understand the evolving nature of the virus. This understanding is vital for implementing effective public health measures to control the spread of the virus and protect public health.
Secondly, studying these sublineages is important for vaccine development. By analyzing the characteristics of the Omicron sublineages, researchers can gain insights into how the virus is evolving and how it may impact the effectiveness of current vaccines. This information can guide the development of updated or new vaccines to ensure continued protection against the virus. Furthermore, understanding the characteristics of these sublineages is crucial for treatment strategies. By monitoring the spread and characteristics of the Omicron sub-lineages, researchers can assess their potential impact on the trajectory of the pandemic and develop targeted treatment approaches to mitigate their effects. Table 1 summarizes key information about the analysed Omicron sub-lineages.
| Table 1: Overview of the Omicron sub-lineages. | |
| Sublineage | Key Information |
| BA.1 | First identified in Botswana, contains the H69/V70 deletion, associated with increased transmissibility. |
| BA.2 | Contains the S371L mutation, potentially more transmissible than BA.1. |
| BA.3 | Contains the S371F mutation, may have increased transmissibility and potential immune evasion. |
| BA.4 | Contains the S371L and S477N mutations, potentially impacting transmissibility and immune evasion. |
| BA.5 | Contains the S371L, S477N, and N679K mutations, potential impact on transmissibility and immune evasion. |
| XBB | Contains the S371L and S477N mutations, similar to BA.4, but with additional mutations. |
| BQ.1.18 | Contains the S371L and S477N mutations, similar to BA.4, but with additional mutations. |
| BA.2.75 | Contains the S371L mutation, similar to BA.2, but with additional mutations. |
| Alpha | First identified in the UK, contains the N501Y mutation, associated with increased transmissibility. |
| Beta | First identified in South Africa, contains the E484K mutation, potentially impact on vaccine efficacy. |
| Gamma | First identified in Brazil, contains the E484K and N501Y mutations, potentially impact on vaccine efficacy. |
| Delta | First identified in India, contains the L452R and P681R mutations, associated with increased transmissibility. |
| B.1.1.529 | Contains the N501Y, E484A, and Q498R mutations, potentially impact on transmissibility and immune evasion. |
| EG.5.1 | Contains the S371L and S477N mutations, similar to BA.4, but with additional mutations. |
It’s important to note that ongoing research is being conducted to understand the implications of these sublineages in terms of transmissibility, severity, and vaccine effectiveness. The situation is evolving, and health authorities continue to monitor these sublineages closely to adapt public health measures and inform vaccine development and treatment strategies.
Finally, monitoring and studying the Omicron sublineages is essential for understanding the evolving nature of the virus, and informing public health measures, vaccine development, and treatment strategies. This ongoing research is crucial for addressing the potential impact of the Omicron variant on the trajectory of the pandemic and ensuring the continued effectiveness of public health responses.
Retrieve data on new Omicron subtypes to study how they are related and what mutations they have
We gathered information about the new types of the Omicron variant from various sources like scientific databases, the World Health Organization [33-35], and the CDC [36]. We used different keywords to search for this information such as “sublineages of Omicron”,” B.1.1.529”, “BA.1”, “BA.2”, “BA.4”, “BA.5”, “BA.2.12.1” and “BA.2.75”.. We also looked at data from the GISAID database [37,38] and obtained the spike protein’s PDB file from the RCSB-PDB database [39].
Studying new Omicron subtypes to see how they are related to each other at a molecular level
We studied different types of the Omicron variant, like BA.1, BA.2, BA.4, BA.5, BA.2.12.1, and BA.2.75. We used the Nextstrain server, which uses the GISAID database, to analyze how these variants are related to each other [40,41]. We focused on Omicron (B.1.1.529 or Nextstrain clade: 21M) and its subtypes, such as BA.1 (Nextstrain clade: 21K), BA.2 (Nextstrain clade: 21L), BA.4 (Nextstrain clade: 22A), BA.5 (Nextstrain clade: 22B), BA.2.12.1 (Nextstrain clade: 22C), and BA. 2.75 (Nextstrain clade: 22D) for our study.
Studying new Omicron subtypes to see how they are changing
We gathered data on Omicron mutations and their sublineages from the CoVariant server [42]. However, we didn’t look at all the mutations to study how often risky mutations occur and their characteristics.
We used databases from CNCB-NGDC and other coronavirus resources to create a heatmap showing mutations in the spike protein of Omicron sublineages [43,44]. The VarEPS server helped us identify important mutations related to risk and how well neutralizing antibodies bind to the spike protein [45]. We also used the DynaMut server to analyze changes in the stability of the S-glycoprotein. We looked at the ΔΔG score through simulation on a server [46]. To create graphs, plots, and models [47], we used the PAST statistical software and MATLAB [48].
So far, the pipelines we’ve built have allowed us to apply a fair and accurate automated way to determine how dangerous SARS-CoV-2 is in hamsters. We’re now utilising this strategy to see how dangerous the newly discovered omicron sublineages are.
The BA.5 sublineage of the Omicron variant has been a topic of concern due to its potential impact on public health. Virulence refers to the severity of the disease caused by a particular virus. Assessing the virulence of BA.5 involves examining various factors, including its ability to cause severe illness, hospitalization rates, and mortality. Preliminary data suggests that BA.5 may have increased transmissibility compared to earlier variants, but its impact on disease severity is still being studied. It’s important to consider that virulence is just one aspect of a virus’s overall impact. Factors such as vaccination status, healthcare infrastructure, and public health measures also play a crucial role in determining the overall impact of a variant. Ongoing research and surveillance are essential to better understand the virulence of BA.5 and its implications for public health. It’s crucial to monitor data from affected regions, conduct clinical studies, and analyze real-world outcomes to inform public health strategies and interventions. This will help in developing targeted responses to mitigate the impact of BA.5 and other emerging variants.
The virulence of the XBB Omicron sublineage is currently a topic of intense research and discussion within the scientific community. Virulence refers to the severity of the disease caused by a particular virus, including factors such as its ability to cause illness and the potential for severe outcomes. As of the current date, there is ongoing research to understand the impact of the XBB Omicron sublineage on disease severity, transmissibility, and immune evasion. Preliminary data suggests that this sublineage may have mutations that could potentially impact its behaviour, but further studies are needed to fully assess its virulence. It’s important to note that assessing virulence is a complex process that involves studying various factors, including clinical outcomes, epidemiological data, and laboratory experiments. Researchers are working diligently to gather and analyze data to better understand the implications of the XBB Omicron sub-lineage. As more information becomes available, it will be crucial to closely monitor the scientific literature and official updates from public health authorities for the latest assessments of the virulence of the XBB Omicron sublineage.
The B.1.1.529 Omicron variant, also known as B.1.1.529.1 or BA.1.1, has been a topic of significant discussion and concern within the scientific and public health communities. This variant, first identified in Botswana and South Africa, has raised questions about its potential impact on public health due to its large number of mutations, particularly within the spike protein. The virulence of the B.1.1.529 Omicron variant is currently being closely studied, and it is important to note that our understanding of its characteristics is evolving as new data becomes available. Initial reports suggest that the Omicron variant may have an increased ability to evade immunity, potentially impacting the effectiveness of existing vaccines and previous infection-induced immunity. However, the severity of illness caused by the Omicron variant is still being investigated, and it is not yet clear whether it leads to more severe disease compared to other variants. It is crucial for researchers and public health authorities to continue monitoring the spread and impact of the Omicron variant, as well as conducting in-depth studies to assess its virulence, transmissibility, and potential impact on vaccine effectiveness. This ongoing research will be essential for informing public health measures and guiding the development of targeted strategies to mitigate the impact of the Omicron variant on global health.
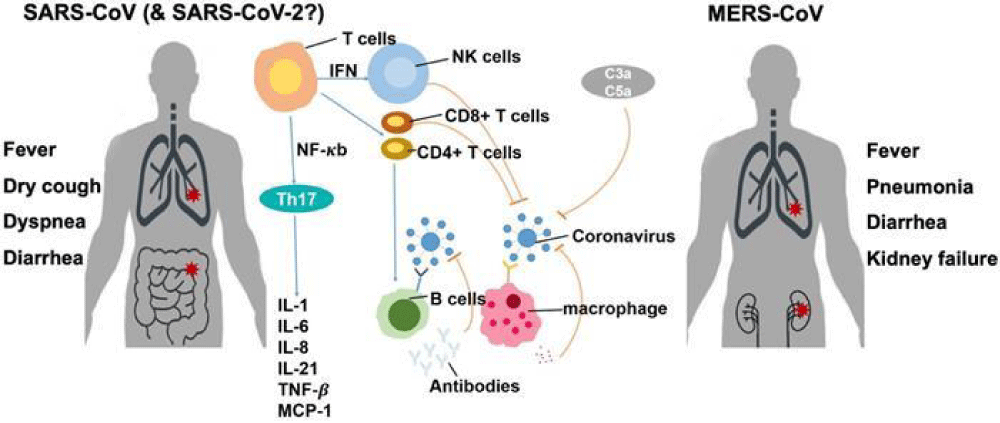
Figure 3 shows how different versions of the SARS-CoV-2 virus affect our immune system and how easily the disease spreads. The changes in the spike protein of the Omicron variant of SARS-CoV-2 make it better at infecting people by sticking more strongly to their ACE2 receptors. This also makes it harder for antibodies from vaccines or previous infections to stop the virus. These changes help the virus spread more easily among both vaccinated and unvaccinated people.
Figure 3: SARS-CoV-2 Variants and Immune Response [61].
The BA.2 and BA.2.75 sublineages of the Omicron variant have been of particular interest due to their potential impact on public health. These sublineages have been reported to exhibit increased transmissibility compared to the original Omicron variant (BA.1). However, it’s important to note that increased transmissibility does not necessarily equate to increased virulence. Virulence refers to the severity of the disease caused by a particular virus. While the BA.2 and BA.2.75 sublineages may be more transmissible, it’s crucial to conduct thorough studies to determine whether they also cause more severe illness or lead to higher rates of hospitalization and mortality compared to the original Omicron variant or other variants of concern.
Assessing the virulence of these sublineages requires comprehensive clinical data analysis, including monitoring the severity of illness in infected individuals, hospitalization rates, and mortality rates. Additionally, laboratory studies can help determine whether these sub-lineages exhibit any differences in their ability to cause severe disease or evade the immune response compared to other variants. It’s also important to consider the impact of vaccination and prior immunity on the virulence of these sublineages. Understanding how these sub-lineages interact with the immune system and whether they are more or less susceptible to vaccine-induced immunity is crucial in assessing their overall impact on public health. In-depth discussions and analyses by public health authorities, epidemiologists, virologists, and other experts are essential to monitor and assess the virulence of these sublineages and to inform public health measures and interventions. Ongoing surveillance and research will be critical in understanding the full implications of these Omicron sublineages on global health.
Figure 4 shows the different versions of the Omicron variant, their mutations, and how they are more infectious and easily spread. The Omicron variant can infect both adults and children and mainly affects the respiratory system, intestines, and brain. It also changes to better attach to human cells, making it easier for the virus to spread and infect people.
Figure 4: Omicron Variant: Mutations and Infectiousness [62].
Studying how easily the virus spreads, how sick it makes people, how well tests work, how good vaccines are, and how well treatments work will help us deal with the Omicron variant and other new versions of the virus. You can find a summary of the plans to deal with these new variants in Figure 5.
Figure 5: Methods for preventing SARS-CoV-2 developing mutations [63].
The virulence of the EG.5.1 Omicron sublineage is currently being closely studied by researchers and public health officials. Virulence refers to the severity of the disease caused by a particular virus strain. In the case of EG.5.1 Omicron, initial reports suggest that it may have increased transmissibility compared to previous variants, but its impact on disease severity is still being investigated. Several factors are considered when assessing the virulence of a virus, including its ability to cause severe illness, hospitalization rates, and mortality. Researchers are conducting studies to determine whether EG.5.1 Omicron leads to more severe disease outcomes compared to other variants, such as Delta or the original strain of the virus. It’s important to note that assessing virulence is a complex process that requires comprehensive data analysis and clinical observations. As more information becomes available, public health authorities will be able to provide a more detailed assessment of the virulence of EG.5.1 Omicron and its implications for public health measures.
Throughout the COVID-19 pandemic, new variants of the virus have been appearing periodically, and some of these variants may be different in how harmful they are compared to earlier ones. It’s important to keep a close eye on the harmfulness of any new variant as it emerges. Scientists have been using animal models to learn more about how the virus causes disease [49]. So far, there has been a strong connection between how harmful variants like Delta and BA.1 are in humans and how they affect hamsters that have been infected in experiments [50,51].
The research we mentioned about using body weight loss, lung function, and lung tissue features in hamsters to assess the strength of different SARS-CoV-2 virus variants is quite interesting [52,53]. It’s not uncommon for scientists to use animal models to study infectious diseases, as they can provide valuable insights into how the virus affects the body and how different variants compare in terms of virulence. Body weight loss in animal models can be a useful indicator of the severity of the infection. A more virulent virus variant is likely to cause greater weight loss in infected animals compared to a less virulent one. Lung function and lung tissue features are also crucial parameters to consider, as they directly reflect the impact of the virus on the respiratory system. Variants that cause more severe lung damage and compromise lung function are generally considered to be stronger or more virulent.
The fact that these measurements have been good indicators of the strength of the virus, particularly in the case of variants like Delta and Omicron, highlights the importance of understanding the biological impact of different viral strains. The differences in strength between variants, especially those with very different levels of strength like Delta and Omicron, can have significant implications for public health and the development of targeted interventions. However, it’s also important to acknowledge the potential challenges in comparing the strength of variants when the differences are small, such as with BA.2 and BA.5 compared to BA.1. Variants that are closely related may exhibit subtle differences in their impact on the host, making it more challenging to discern their relative strength. The discrepancies between studies in such cases, as mentioned in references [54,55], underscore the complexity of assessing the strength of closely related variants and the need for rigorous and consistent methodologies in research.
Overall, the use of animal models to study the strength of SARS-CoV-2 variants provides valuable insights into the biological differences between these strains. It also emphasizes the importance of considering multiple parameters, such as body weight loss, lung function, and lung tissue features, to comprehensively evaluate the virulence of different variants. This research is crucial for understanding the potential impact of emerging variants on public health and for informing strategies to mitigate their spread and severity.
The study demonstrates that certain variants, specifically BA.5, BA.2.75, and EG.5.1, have developed more harmful characteristics compared to the BA.1 variant. This conclusion is supported by other recent studies referenced as [54,56]. Additionally, the study found no observed differences in harmfulness between XBB and BA.1, which aligns with another recent study [57]. Furthermore, the study noted that BA.2 tended to cause more macrophage infiltrates and type 2 pneumocyte hyperplasia compared to BA.1, although these differences were not statistically significant. This observation is consistent with other studies that found no major differences in harmfulness between BA.1 and BA.2 [57,58]. However, it contrasts with a previous study that used recombinant viruses with either BA.1 or BA.2 spike [58].
The findings we have mentioned are indeed crucial in our ongoing battle against the SARS-CoV-2 virus. The identification of different variants with varying levels of harmfulness and characteristics underscores the need for continuous monitoring and research to understand how the virus is evolving and its potential impact on public health. Firstly, the fact that certain variants exhibit more harmful characteristics emphasizes the importance of ongoing surveillance. By closely monitoring the genetic makeup and behaviour of the virus, researchers and public health officials can better understand how it is changing over time. This surveillance allows for early detection of concerning variants and informs public health responses to mitigate their spread and impact.
Furthermore, these findings highlight the necessity for continued vigilance in developing and updating public health measures and interventions. As the virus evolves, so too must our strategies for controlling its spread and mitigating its effects. This may involve adapting vaccination strategies, updating diagnostic tests, and refining treatment protocols to address the specific characteristics of emerging variants. In addition, the significance of these findings underscores the need for global collaboration and information sharing. The evolution of the virus knows no borders, and a coordinated international effort is essential to track and respond to new variants effectively. This includes sharing genomic data, research findings, and best practices in public health interventions to ensure a unified and informed response to the evolving nature of the virus.
Finally, the identification of different SARS-CoV-2 variants with varying harmful characteristics emphasizes the critical importance of ongoing surveillance, research, and global cooperation. By staying vigilant and proactive in our approach, we can better understand and address the evolving nature of the virus, ultimately working towards minimizing its impact on public health.
The emergence of newly identified Omicron sublineages has raised concerns about potential limitations in our understanding of the virus and its variants. To address these limitations and provide insights into future research directions, it is crucial to consider several key aspects. Firstly, genomic surveillance and sequencing efforts need to be intensified to track the evolution of the virus and identify new sublineages. This will help in understanding the genetic diversity of the virus and its implications for transmissibility, severity, and vaccine efficacy. Additionally, it will aid in the early detection of potentially concerning variants. Secondly, comprehensive epidemiological studies are essential to assess the impact of these sublineages on transmission dynamics, disease severity, and immune escape. Understanding the clinical characteristics and outcomes associated with specific sublineages is critical for informing public health interventions and treatment strategies.
Furthermore, laboratory-based research is needed to investigate the antigenic and functional properties of the newly identified sublineages. This includes assessing their susceptibility to neutralization by antibodies elicited through natural infection or vaccination. Such studies will provide valuable insights into the potential impact of these sublineages on vaccine effectiveness and the development of future vaccine candidates. In addition, it is important to conduct studies to evaluate the effectiveness of existing diagnostic tests, therapeutics, and preventive measures against these sublineages. This will help in optimizing strategies for disease detection, management, and control in the context of evolving viral diversity.
Moreover, collaborative efforts at the global level are crucial to facilitate data sharing, research coordination, and the development of standardized approaches for characterizing and monitoring Omicron sublineages. This will enable a more comprehensive and timely understanding of the evolving landscape of SARS-CoV-2 variants. Finally, addressing the potential limitations associated with newly identified Omicron sublineages requires a multidisciplinary approach encompassing genomic surveillance, epidemiological studies, laboratory research, and global collaboration. By focusing on these areas, we can gain valuable insights into the characteristics of these sublineages and inform evidence-based public health responses and future research directions.
The new Omicron variant of COVID-19 is causing a big increase in cases around the world. It’s also making vaccines and treatments less effective. We need to make sure everyone can get vaccines and that we make more of them. We also need to keep giving out vaccines to as many people as possible. To fight the new variant and stop it from spreading, we should wear masks, wash our hands, stay apart from others, keep things clean, and make sure our hospitals are ready. We also need to plan for future pandemics.
We need to do more research to understand why the new variants of the virus that cause COVID-19 spread more easily, how they are changing over time, and how they affect people. We also need to figure out how well previous infections protect people and how the virus can avoid the immune system. We should make better vaccines, give extra vaccine doses, and create new vaccines that work against the new variants. We also need to develop new treatments and drugs to help people who get sick with COVID-19. These improvements will help us deal with the new variants of the virus that are causing more infections and making people sicker.
We still have a lot of questions about the Omicron variant and its different types. We need to keep a close watch on it and other new versions of the virus. To make vaccines and treatments, we can focus on certain parts of the virus, make vaccines that work against many versions of the virus, and use strong additives to make the vaccines more effective. This could help protect against the current Omicron variant and any new versions, as well as any future viruses like SARS-CoV-3.
- Carabelli AM, Peacock TP, Thorne LG, Harvey WT, Hughes J; COVID-19 Genomics UK Consortium; Peacock SJ, Barclay WS, de Silva TI, Towers GJ, Robertson DL. SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nat Rev Microbiol. 2023 Mar;21(3):162-177. doi: 10.1038/s41579-022-00841-7. Epub 2023 Jan 18. PMID: 36653446; PMCID: PMC9847462.
- Tracking SARS-CoV-2 Variants. 2022. https://www.who.int/activities/tracking-SARS-CoV-2- variants.
- Tegally H, Moir M, Everatt J, Giovanetti M, Scheepers C, Wilkinson E, Subramoney K, Makatini Z, Moyo S, Amoako DG, Baxter C, Althaus CL, Anyaneji UJ, Kekana D, Viana R, Giandhari J, Lessells RJ, Maponga T, Maruapula D, Choga W, Matshaba M, Mbulawa MB, et al. Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nat Med. 2022 Sep;28(9):1785-1790. doi: 10.1038/s41591-022-01911-2. Epub 2022 Jun 27. PMID: 35760080; PMCID: PMC9499863.
- Viana R, Moyo S, Amoako DG, Tegally H, Scheepers C, Althaus CL, Anyaneji UJ, Bester PA, Boni MF, Chand M, Choga WT, Colquhoun R, Davids M, Deforche K, Doolabh D, du Plessis L, Engelbrecht S, Everatt J, Giandhari J, Giovanetti M, Hardie D, Hill V, Hsiao NY, Iranzadeh A, Ismail A, Joseph C, Joseph R, Koopile L, Kosakovsky Pond SL, et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022 Mar;603(7902):679-686. doi: 10.1038/s41586-022-04411-y. Epub 2022 Jan 7. PMID: 35042229; PMCID: PMC8942855.
- Cherian S, Potdar V, Jadhav S, Yadav P, Gupta N, Das M, Rakshit P, Singh S, Abraham P, Panda S, Team N. SARS-CoV-2 Spike Mutations, L452R, T478K, E484Q and P681R, in the Second Wave of COVID-19 in Maharashtra, India. Microorganisms. 2021 Jul 20;9(7):1542. doi: 10.3390/microorganisms9071542. PMID: 34361977; PMCID: PMC8307577.
- Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, Pearson CAB, Russell TW, Tully DC, Washburne AD, Wenseleers T, Gimma A, Waites W, Wong KLM, van Zandvoort K, Silverman JD; CMMID COVID-19 Working Group; COVID-19 Genomics UK (COG-UK) Consortium; Diaz-Ordaz K, Keogh R, Eggo RM, Funk S, Jit M, Atkins KE, Edmunds WJ. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021 Apr 9;372(6538):eabg3055. doi: 10.1126/science.abg3055. Epub 2021 Mar 3. PMID: 33658326; PMCID: PMC8128288.
- Tegally H, Moir M, Everatt J, Giovanetti M, Scheepers C, Wilkinson E, de Oliveira T. Continued emergence and evolution of Omicron in South Africa: New BA. 4 and BA. 5 lineages. MedRxiv. 2022; 2022-05.
- Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, Hinsley WR, Laydon DJ, Dabrera G, O'Toole Á, Amato R, Ragonnet-Cronin M, Harrison I, Jackson B, Ariani CV, Boyd O, Loman NJ, McCrone JT, Gonçalves S, Jorgensen D, Myers R, Hill V, Jackson DK, Gaythorpe K, Groves N, Sillitoe J, Kwiatkowski DP; COVID-19 Genomics UK (COG-UK) consortium; Flaxman S, Ratmann O, Bhatt S, Hopkins S, Gandy A, Rambaut A, Ferguson NM. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021 May;593(7858):266-269. doi: 10.1038/s41586-021-03470-x. Epub 2021 Mar 25. PMID: 33767447.
- Callaway E. What Omicron's BA.4 and BA.5 variants mean for the pandemic. Nature. 2022 Jun;606(7916):848-849. doi: 10.1038/d41586-022-01730-y. PMID: 35750920.
- Brüssow H. COVID-19: Omicron - the latest, the least virulent, but probably not the last variant of concern of SARS-CoV-2. Microb Biotechnol. 2022 Jul;15(7):1927-1939. doi: 10.1111/1751-7915.14064. Epub 2022 Apr 20. PMID: 35443078; PMCID: PMC9111164.
- Dhama K, Nainu F, Frediansyah A, Yatoo MI, Mohapatra RK, Chakraborty S, Zhou H, Islam MR, Mamada SS, Kusuma HI, Rabaan AA, Alhumaid S, Mutair AA, Iqhrammullah M, Al-Tawfiq JA, Mohaini MA, Alsalman AJ, Tuli HS, Chakraborty C, Harapan H. Global emerging Omicron variant of SARS-CoV-2: Impacts, challenges and strategies. J Infect Public Health. 2023 Jan;16(1):4-14. doi: 10.1016/j.jiph.2022.11.024. Epub 2022 Nov 19. PMID: 36446204; PMCID: PMC9675435.
- Callaway E. Heavily mutated Omicron variant puts scientists on alert. Nature. 2021 Dec;600(7887):21. doi: 10.1038/d41586-021-03552-w. PMID: 34824381.
- Taylor CA, Whitaker M, Anglin O, Milucky J, Patel K, Pham H, Chai SJ, Alden NB, Yousey-Hindes K, Anderson EJ, Teno K, Reeg L, Como-Sabetti K, Bleecker M, Barney G, Bennett NM, Billing LM, Sutton M, Talbot HK, McCaffrey K, Havers FP; COVID-NET Surveillance Team. COVID-19-Associated Hospitalizations Among Adults During SARS-CoV-2 Delta and Omicron Variant Predominance, by Race/Ethnicity and Vaccination Status - COVID-NET, 14 States, July 2021-January 2022. MMWR Morb Mortal Wkly Rep. 2022 Mar 25;71(12):466-473. doi: 10.15585/mmwr.mm7112e2. PMID: 35324880; PMCID: PMC8956338.
- Mohapatra RK, Kandi V, Sarangi AK, Verma S, Tuli HS, Chakraborty S, Chakraborty C, Dhama K. The recently emerged BA.4 and BA.5 lineages of Omicron and their global health concerns amid the ongoing wave of COVID-19 pandemic - Correspondence. Int J Surg. 2022 Jul;103:106698. doi: 10.1016/j.ijsu.2022.106698. Epub 2022 Jun 8. PMID: 35690362; PMCID: PMC9176102.
- Fan Y, Li X, Zhang L, Wan S, Zhang L, Zhou F. SARS-CoV-2 Omicron variant: recent progress and future perspectives. Signal Transduct Target Ther. 2022 Apr 28;7(1):141. doi: 10.1038/s41392-022-00997-x. PMID: 35484110; PMCID: PMC9047469.
- Callaway E. Are COVID surges becoming more predictable? New Omicron variants offer a hint. Nature. 2022 May;605(7909):204-206. doi: 10.1038/d41586-022-01240-x. PMID: 35523871.
- Cao Y, Wang J, Jian F, Xiao T, Song W, Yisimayi A, Huang W, Li Q, Wang P, An R, Wang J, Wang Y, Niu X, Yang S, Liang H, Sun H, Li T, Yu Y, Cui Q, Liu S, Yang X, Du S, Zhang Z, Hao X, Shao F, Jin R, Wang X, Xiao J, Wang Y, Xie XS. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022 Feb;602(7898):657-663. doi: 10.1038/s41586-021-04385-3. Epub 2021 Dec 23. PMID: 35016194; PMCID: PMC8866119.
- Hoffmann M, Krüger N, Schulz S, Cossmann A, Rocha C, Kempf A, Nehlmeier I, Graichen L, Moldenhauer AS, Winkler MS, Lier M, Dopfer-Jablonka A, Jäck HM, Behrens GMN, Pöhlmann S. The Omicron variant is highly resistant against antibody-mediated neutralization: Implications for control of the COVID-19 pandemic. Cell. 2022 Feb 3;185(3):447-456.e11. doi: 10.1016/j.cell.2021.12.032. Epub 2021 Dec 24. PMID: 35026151; PMCID: PMC8702401.
- Wilhelm A, Widera M, Grikscheit K, Toptan T, Schenk B, Pallas C, Metzler M, Kohmer N, Hoehl S, Marschalek R, Herrmann E, Helfritz FA, Wolf T, Goetsch U, Ciesek S. Limited neutralisation of the SARS-CoV-2 Omicron subvariants BA.1 and BA.2 by convalescent and vaccine serum and monoclonal antibodies. EBioMedicine. 2022 Aug;82:104158. doi: 10.1016/j.ebiom.2022.104158. Epub 2022 Jul 11. PMID: 35834885; PMCID: PMC9271884.
- Carrazco-Montalvo A, Herrera-Yela A, Alarcón-Vallejo D, Gutiérrez-Pallo D, Armendáriz-Castillo I, Andrade-Molina D, Muñoz-Mawyin K, Fernández-Cadena JC, Morey-León G, Usfq-Covid-Consortium, Crn Influenza Y Ovr-Inspi, Patiño L. Omicron Sub-Lineages (BA.1.1.529 + BA.*) Current Status in Ecuador. Viruses. 2022 May 28;14(6):1177. doi: 10.3390/v14061177. PMID: 35746651; PMCID: PMC9230377.
- Kumar S, Karuppanan K, Subramaniam G. Omicron (BA.1) and sub-variants (BA.1.1, BA.2, and BA.3) of SARS-CoV-2 spike infectivity and pathogenicity: A comparative sequence and structural-based computational assessment. J Med Virol. 2022 Oct;94(10):4780-4791. doi: 10.1002/jmv.27927. Epub 2022 Jun 16. PMID: 35680610; PMCID: PMC9347785.
- Desingu PA, Nagarajan K, Dhama K. Emergence of Omicron third lineage BA.3 and its importance. J Med Virol. 2022 May;94(5):1808-1810. doi: 10.1002/jmv.27601. Epub 2022 Jan 23. PMID: 35043399; PMCID: PMC9015590.
- Mohapatra RK, Kandi V, Verma S, Dhama K. Challenges of the Omicron (B.1.1.529) Variant and Its Lineages: A Global Perspective. Chembiochem. 2022 May 4;23(9):e202200059. doi: 10.1002/cbic.202200059. Epub 2022 Mar 23. PMID: 35322516; PMCID: PMC9083815.
- Khandia R, Singhal S, Alqahtani T, Kamal MA, El-Shall NA, Nainu F, Desingu PA, Dhama K. Emergence of SARS-CoV-2 Omicron (B.1.1.529) variant, salient features, high global health concerns and strategies to counter it amid ongoing COVID-19 pandemic. Environ Res. 2022 Jun;209:112816. doi: 10.1016/j.envres.2022.112816. Epub 2022 Jan 29. PMID: 35093310; PMCID: PMC8798788.
- Tegally H, Moir M, Everatt J, Giovanetti M, Scheepers C, Wilkinson E, Subramoney K, Makatini Z, Moyo S, Amoako DG, Baxter C, Althaus CL, Anyaneji UJ, Kekana D, Viana R, Giandhari J, Lessells RJ, Maponga T, Maruapula D, Choga W, Matshaba M, Mbulawa MB, Msomi N; NGS-SA consortium; Naidoo Y, Pillay S, Sanko TJ, et al. Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nat Med. 2022 Sep;28(9):1785-1790. doi: 10.1038/s41591-022-01911-2. Epub 2022 Jun 27. PMID: 35760080; PMCID: PMC9499863.
- Rahman S, Hossain MJ, Nahar Z, Shahriar M, Bhuiyan MA, Islam MR. Emerging SARS-CoV-2 Variants and Subvariants: Challenges and Opportunities in the Context of COVID-19 Pandemic. Environ Health Insights. 2022 Oct 20;16:11786302221129396. doi: 10.1177/11786302221129396. PMID: 36299441; PMCID: PMC9585367.
- Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, Du S, Wang J, Li Q, Chen X, Yu Y, Wang P, Zhang Z, Liu P, An R, Hao X, Wang Y, Wang J, Feng R, Sun H, Zhao L, Zhang W, Zhao D, Zheng J, Yu L, Li C, Zhang N, Wang R, Niu X, Yang S, Song X, Chai Y, Hu Y, Shi Y, Zheng L, Li Z, Gu Q, Shao F, Huang W, Jin R, Shen Z, Wang Y, Wang X, Xiao J, Xie XS. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022 Aug;608(7923):593-602. doi: 10.1038/s41586-022-04980-y. Epub 2022 Jun 17. PMID: 35714668; PMCID: PMC9385493.
- Basky G, Vogel L. XE, XD & XF: what to know about the Omicron hybrid variants. CMAJ. 2022 May 9;194(18):E654-E655. doi: 10.1503/cmaj.1095998. PMID: 35534032; PMCID: PMC9259411.
- Chakraborty C, Bhattacharya M, Sharma AR, Dhama K. Recombinant SARS-CoV-2 variants XD, XE, and XF: The emergence of recombinant variants requires an urgent call for research - Correspondence. Int J Surg. 2022 Jun;102:106670. doi: 10.1016/j.ijsu.2022.106670. Epub 2022 May 13. PMID: 35569759; PMCID: PMC9098807.
- Mohapatra RK, Kandi V, Tuli HS, Chakraborty C, Dhama K. The recombinant variants of SARS-CoV-2: Concerns continues amid COVID-19 pandemic. J Med Virol. 2022 Aug;94(8):3506-3508. doi: 10.1002/jmv.27780. Epub 2022 Apr 27. PMID: 35419806; PMCID: PMC9088633.
- Rahimi F, Talebi Bezmin Abadi A. Hybrid SARS-CoV-2 variants. Int J Surg. 2022 Jun;102:106656. doi: 10.1016/j.ijsu.2022.106656. Epub 2022 May 6. PMID: 35533853; PMCID: PMC9074379.
- Roemer C, Hisner R, Frohberg N, Sakaguchi H, Gueli F, Peacock TP. SARS-CoV-2 Evolution, Post-Omicron. virological. org. 911.
- WHO: Enhancing readiness for Omicron (B.1.1.529): technical brief and priority actions for member states. 2022. https://www.who.int/docs/default-source/coronaviruse/technical-brief-and-priority-action-on-omicron.pdf?sfvrsn=50732953_3
- WHO, South Africa. 2021, 2022. https://covid19.who.int/region/afro/country/za
- WHO, Statement on Omicron sublineage BA.2, 2022. https://www.who.int/news/item/22–02-2022-statement-on-omicron-sublineage-ba.2
- CDC, Omicron variant: what you need to know. 2021, 2022. https://www.cdc.gov/coronavirus/2019-ncov/variants/omicron-variant.html
- Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017 Mar 30;22(13):30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. PMID: 28382917; PMCID: PMC5388101.
- Elbe S, Buckland-Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob Chall. 2017 Jan 10;1(1):33-46. doi: 10.1002/gch2.1018. PMID: 31565258; PMCID: PMC6607375.
- Westbrook J, Feng Z, Jain S, Bhat TN, Thanki N, Ravichandran V, Gilliland GL, Bluhm W, Weissig H, Greer DS, Bourne PE, Berman HM. The Protein Data Bank: unifying the archive. Nucleic Acids Res. 2002 Jan 1;30(1):245-8. doi: 10.1093/nar/30.1.245. PMID: 11752306; PMCID: PMC99110.
- Nextstrain: Nextstrain SARS-CoV-2 Resources on Nextstrain. 2021. https://nextstrain.org/sars-cov-2/
- Mathieu E, Ritchie H, Ortiz-Ospina E, Roser M, Hasell J, Appel C, Giattino C, Rodés-Guirao L. A global database of COVID-19 vaccinations. Nat Hum Behav. 2021 Jul;5(7):947-953. doi: 10.1038/s41562-021-01122-8. Epub 2021 May 10. Erratum in: Nat Hum Behav. 2021 Jun 17;: PMID: 33972767.
- HE. Coresearchers, CoVariants. 2021. https://covariants.org.
- CNCB-NGDC Members and Partners. Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2022. Nucleic Acids Res. 2022 Jan 7;50(D1):D27-D38. doi: 10.1093/nar/gkab951. PMID: 34718731; PMCID: PMC8728233.
- Zhao WM, Song SH, Chen ML, Zou D, Ma LN, Ma YK, Li RJ, Hao LL, Li CP, Tian DM, Tang BX, Wang YQ, Zhu JW, Chen HX, Zhang Z, Xue YB, Bao YM. The 2019 novel coronavirus resource. Yi Chuan. 2020 Feb 20;42(2):212-221. doi: 10.16288/j.yczz.20-030. PMID: 32102777.
- Sun Q, Shu C, Shi W, Luo Y, Fan G, Nie J, Bi Y, Wang Q, Qi J, Lu J, Zhou Y, Shen Z, Meng Z, Zhang X, Yu Z, Gao S, Wu L, Ma J, Hu S. VarEPS: an evaluation and prewarning system of known and virtual variations of SARS-CoV-2 genomes. Nucleic Acids Res. 2022 Jan 7;50(D1):D888-D897. doi: 10.1093/nar/gkab921. PMID: 34634813; PMCID: PMC8728250.
- Rodrigues CH, Pires DE, Ascher DB. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018 Jul 2;46(W1):W350-W355. doi: 10.1093/nar/gky300. PMID: 29718330; PMCID: PMC6031064.
- Hammer Ø, Harper DA. Past: paleontological statistics software package for education and data analysis. Palaeontologia electronica. 2001; 4(1): 1.
- MathWorks Inc. MATLAB, High-performance Numeric Computation, and Visualization Software: User's Guide: for UNIX Workstations. MathWorks. 1992.
- Imai M, Iwatsuki-Horimoto K, Hatta M, Loeber S, Halfmann PJ, Nakajima N, Watanabe T, Ujie M, Takahashi K, Ito M, Yamada S, Fan S, Chiba S, Kuroda M, Guan L, Takada K, Armbrust T, Balogh A, Furusawa Y, Okuda M, Ueki H, et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Natl Acad Sci U S A. 2020 Jul 14;117(28):16587-16595. doi: 10.1073/pnas.2009799117. Epub 2020 Jun 22. PMID: 32571934; PMCID: PMC7368255.
- Chan JF, Zhang AJ, Yuan S, Poon VK, Chan CC, Lee AC, Chan WM, Fan Z, Tsoi HW, Wen L, Liang R, Cao J, Chen Y, Tang K, Luo C, Cai JP, Kok KH, Chu H, Chan KH, Sridhar S, Chen Z, Chen H, To KK, Yuen KY. Simulation of the Clinical and Pathological Manifestations of Coronavirus Disease 2019 (COVID-19) in a Golden Syrian Hamster Model: Implications for Disease Pathogenesis and Transmissibility. Clin Infect Dis. 2020 Dec 3;71(9):2428-2446. doi: 10.1093/cid/ciaa325. PMID: 32215622; PMCID: PMC7184405.
- Suzuki R, Yamasoba D, Kimura I, Wang L, Kishimoto M, Ito J, Morioka Y, Nao N, Nasser H, Uriu K, Kosugi Y, Tsuda M, Orba Y, Sasaki M, Shimizu R, Kawabata R, Yoshimatsu K, Asakura H, Nagashima M, Sadamasu K, Yoshimura K; Genotype to Phenotype Japan (G2P-Japan) Consortium; Sawa H, Ikeda T, Irie T, Matsuno K, Tanaka S, Fukuhara T, Sato K. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature. 2022 Mar;603(7902):700-705. doi: 10.1038/s41586-022-04462-1. Epub 2022 Feb 1. PMID: 35104835; PMCID: PMC8942852.
- Armando F, Beythien G, Kaiser FK, Allnoch L, Heydemann L, Rosiak M, Becker S, Gonzalez-Hernandez M, Lamers MM, Haagmans BL, Guilfoyle K, van Amerongen G, Ciurkiewicz M, Osterhaus ADME, Baumgärtner W. SARS-CoV-2 Omicron variant causes mild pathology in the upper and lower respiratory tract of hamsters. Nat Commun. 2022 Jun 20;13(1):3519. doi: 10.1038/s41467-022-31200-y. PMID: 35725735; PMCID: PMC9207884.
- Kimura I, Yamasoba D, Tamura T, Nao N, Suzuki T, Oda Y, Mitoma S, Ito J, Nasser H, Zahradnik J, Uriu K, Fujita S, Kosugi Y, Wang L, Tsuda M, Kishimoto M, Ito H, Suzuki R, Shimizu R, Begum MM, Yoshimatsu K, Kimura KT, Sasaki J, Sasaki-Tabata K, Yamamoto Y, Nagamoto T, Kanamune J, Kobiyama K, Asakura H, Nagashima M, Sadamasu K, Yoshimura K, Shirakawa K, Takaori-Kondo A, Kuramochi J, Schreiber G, Ishii KJ; Genotype to Phenotype Japan (G2P-Japan) Consortium; Hashiguchi T, Ikeda T, Saito A, Fukuhara T, Tanaka S, Matsuno K, Sato K. Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5. Cell. 2022 Oct 13;185(21):3992-4007.e16. doi: 10.1016/j.cell.2022.09.018. Epub 2022 Sep 14. PMID: 36198317; PMCID: PMC9472642.
- Uraki R, Halfmann PJ, Iida S, Yamayoshi S, Furusawa Y, Kiso M, Ito M, Iwatsuki-Horimoto K, Mine S, Kuroda M, Maemura T, Sakai-Tagawa Y, Ueki H, Li R, Liu Y, Larson D, Fukushi S, Watanabe S, Maeda K, Pekosz A, Kandeil A, Webby RJ, Wang Z, Imai M, Suzuki T, Kawaoka Y. Characterization of SARS-CoV-2 Omicron BA.4 and BA.5 isolates in rodents. Nature. 2022 Dec;612(7940):540-545. doi: 10.1038/s41586-022-05482-7. Epub 2022 Nov 2. PMID: 36323336.
- Uraki R, Iida S, Halfmann PJ, Yamayoshi S, Hirata Y, Iwatsuki-Horimoto K, Kiso M, Ito M, Furusawa Y, Ueki H, Sakai-Tagawa Y, Kuroda M, Maemura T, Kim T, Mine S, Iwamoto N, Li R, Liu Y, Larson D, Fukushi S, Watanabe S, Maeda K, Wang Z, Ohmagari N, Theiler J, Fischer W, Korber B, Imai M, Suzuki T, Kawaoka Y. Characterization of SARS-CoV-2 Omicron BA.2.75 clinical isolates. Nat Commun. 2023 Mar 23;14(1):1620. doi: 10.1038/s41467-023-37059-x. PMID: 36959194; PMCID: PMC10035475.
- Tamura T, Ito J, Uriu K, Zahradnik J, Kida I, Anraku Y, Nasser H, Shofa M, Oda Y, Lytras S, Nao N, Itakura Y, Deguchi S, Suzuki R, Wang L, Begum MM, Kita S, Yajima H, Sasaki J, Sasaki-Tabata K, Shimizu R, Tsuda M, Kosugi Y, Fujita S, Pan L, Sauter D, Yoshimatsu K, Suzuki S, Asakura H, Nagashima M, Sadamasu K, Yoshimura K, et al. Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants. Nat Commun. 2023 May 16;14(1):2800. doi: 10.1038/s41467-023-38435-3. PMID: 37193706; PMCID: PMC10187524.
- Uraki R, Kiso M, Iida S, Imai M, Takashita E, Kuroda M, Halfmann PJ, Loeber S, Maemura T, Yamayoshi S, Fujisaki S, Wang Z, Ito M, Ujie M, Iwatsuki-Horimoto K, Furusawa Y, Wright R, Chong Z, Ozono S, Yasuhara A, Ueki H, Sakai-Tagawa Y, Li R, Liu Y, Larson D, Koga M, Tsutsumi T, Adachi E, Saito M, Yamamoto S, Hagihara M, Mitamura K, et al. Characterization and antiviral susceptibility of SARS-CoV-2 Omicron BA.2. Nature. 2022 Jul;607(7917):119-127. doi: 10.1038/s41586-022-04856-1. Epub 2022 May 16. PMID: 35576972; PMCID: PMC10579982.
- Yamasoba D, Kimura I, Nasser H, Morioka Y, Nao N, Ito J, Uriu K, Tsuda M, Zahradnik J, Shirakawa K, Suzuki R, Kishimoto M, Kosugi Y, Kobiyama K, Hara T, Toyoda M, Tanaka YL, Butlertanaka EP, Shimizu R, Ito H, Wang L, Oda Y, Orba Y, Sasaki M, Nagata K, Yoshimatsu K, Asakura H, Nagashima M, Sadamasu K, et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2 spike. Cell. 2022 Jun 9;185(12):2103-2115.e19. doi: 10.1016/j.cell.2022.04.035. Epub 2022 May 2. PMID: 35568035; PMCID: PMC9057982.
- Mistry P, Barmania F, Mellet J, Peta K, Strydom A, Viljoen IM, James W, Gordon S, Pepper MS. SARS-CoV-2 Variants, Vaccines, and Host Immunity. Front Immunol. 2022 Jan 3;12:809244. doi: 10.3389/fimmu.2021.809244. PMID: 35046961; PMCID: PMC8761766.
- Wu Y, Long Y, Wang F, Liu W, Wang Y. Emergence of SARS-CoV-2 Omicron variant and strategies for tackling the infection. Immun Inflamm Dis. 2022 Dec;10(12):e733. doi: 10.1002/iid3.733. PMID: 36444634; PMCID: PMC9639460.
- Yi H, Wang J, Wang J, Lu Y, Zhang Y, Peng R, Lu J, Chen Z. The Emergence and Spread of Novel SARS-CoV-2 Variants. Front Public Health. 2021 Aug 2;9:696664. doi: 10.3389/fpubh.2021.696664. PMID: 34409009; PMCID: PMC8364952.
- Yi Y, Lagniton PNP, Ye S, Li E, Xu RH. COVID-19: what has been learned and to be learned about the novel coronavirus disease. Int J Biol Sci. 2020 Mar 15;16(10):1753-1766. doi: 10.7150/ijbs.45134. PMID: 32226295; PMCID: PMC7098028.
- Yin J, Li C, Ye C, Ruan Z, Liang Y, Li Y, Wu J, Luo Z. Advances in the development of therapeutic strategies against COVID-19 and perspectives in the drug design for emerging SARS-CoV-2 variants. Comput Struct Biotechnol J. 2022;20:824-837. doi: 10.1016/j.csbj.2022.01.026. Epub 2022 Jan 31. PMID: 35126885; PMCID: PMC8802458.