More Information
Submitted: September 21, 2021 | Approved: November 09, 2021 | Published: November 10, 2021
How to cite this article: Monti V, Serpenti F, Farina L, Moiraghi ML, Testi MA, et al. An unusual clonal chromosome abnormality der(17)t(11;17)(q24;p13)inv(11)(q13;q23) in a patient with chronic lymphocytic leukemia. Arch Case Rep. 2021; 5: 028-031.
DOI: 10.29328/journal.acr.1001053
Copyright License: © 2021 Monti V, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Der(17); CLL; Pseudocentric chromosome; FISH; Oncology
An unusual clonal chromosome abnormality der(17)t(11;17)(q24;p13)inv(11)(q13;q23) in a patient with chronic lymphocytic leukemia
Valentina Monti1*, Fabio Serpenti2, Lucia Farina2, Maria Luisa Moiraghi1, Maria Adele Testi1 and Giancarlo Pruneri1
1Department of Pathology, National Cancer Institute of Milan, Italy
2Department of Clinical Oncology and Hematology, National Cancer Institute of
Milan, Italy
*Address for Correspondence: Valentina Monti, Department of Pathology, IRCCS National Cancer Institute Foundation, Via Giacomo Venezian, 1, Milan, Italy, 20133, Email: [email protected]
Chronic Lymphocytic Leukemia (CLL) is a common clonal neoplasm of small, mature B-lymphocytes. CLL is a heterogeneous disease with different clinical presentation, response to treatment and survival. This heterogeneity could be explained by the different genetic aberrations in CLL [1]. The most important chromosomal defects correlated to a poor prognosis are 17p13.1 deletion (loss of TP53) or 11q22.3 deletion (loss of ATM) [1,2]. In the era of new drugs in CLL it is mandatory to perform the interphase fluorescence in situ hybridization (FISH) test in order to assess these aberrations, before starting any lines of therapy [3]. Moreover, recent evidence suggests that complex karyotype (CK) with structural chromosomal aberrations identified by using chromosome-banding analysis may be relevant to better define CLL prognosis. It is known that 80% of CLL patients harbor at cytogenetic aberrations [3].
Among these aberrations, a rare translocation der(17)t(11;17)(q24;p13) associate with a paracentric inversion on the long arm of chromosome 11 has been observed. To date, studies reporting the identification of t(11;17)(q24;p13)inv(11)(q13;q23) in CLL patients are lacking. We describe this case as an illustrative unusual clonal cytogenetic example, to stress the need for large repositories of genetic and clinical information to improve our knowledge of mechanisms of CLL development and their prognostic consequences.
B.G., a 73-year–old man, showed up for a clinical check-up at our Institute in September 2014, since he had been diagnosed with B-CLL, Binet stage B. At diagnosis, a bone marrow (BM) aspirate underwent conventional G-banding chromosomal on stimulated CLL cells by EKAMP010M mitogens (ChromoLympho B Proliferation Kit for the in vitro stimulation of human B Lymphocytes) and FISH analysis. Normal karyotype and deletion of ATM gene by FISH technique on chromosome 11q were detected (36% of analyzed nuclei). No other clinically abnormalities, such as 17p or 13q deletion (del) and trisomy 12, were noted. The patient underwent a regular follow-up until April 2017 when progressive bulky lymphadenopathies and a rapid increase in lymphocytosis were detected. Neither anemia nor thrombocytopenia were present. A BM aspirate was performed again and the analysis of immunoglobulin gene heavy chain variable region (IGHV) rearrangement showed a biclonal CLL population with wild-type IGHV in both clones. A normal karyotype and del(17p) was detected in 24 out of 300 nuclei (8%). At that time FISH for 11q deletion was not performed. The patient was in Binet stage B in need of therapy according to the iwCLL criteria, but he was not considered eligible for Ibrutinib due to cardiac comorbidities. Therefore, he received six cycles of Rituximab-Bendamustine until November 2017 achieving a good partial response as for both adenopathies and lymphocytosis. Six months later, he relapsed with progressive lymphadenopathies and therapy with Rituximab-Idelalisib was started in September 2018. At this time, biologic characteristics of the disease were not evaluated. Patient achieved partial response on Idelalisib that was continued until May 2020 when severe drug-related colitis resolved only after drug discontinuation. After six months without therapy, adenopathies progressed again and the patient also developed anemia and thrombocytopenia with a BM CLL infiltrate > 80%.
Del(17p) and del(11q) were detected at FISH analysis on BM leading to TP53 and ATM loss (90% and 85% of analyzed nuclei, respectively) . The TP53 mutation was also present by Next Generation Sequencing (NGS) analysis. Additionally an unusual large clone (> 50% metaphases) emerged with a abnormal karyotype described according to the International System for Human Cytogenetic Nomenclature (ISCN): 45,XY,-11,der(17)t(11;17)(q24;p13)inv(11)(q13;q23)[12/20] (Figure 1).
Figure 1: G-banded Human Kariogram showing the abnormal chromosome 17 marked by star: 45,XY,-11,der(17)t(11;17)(q24;p13)inv(11)(q13;q23).
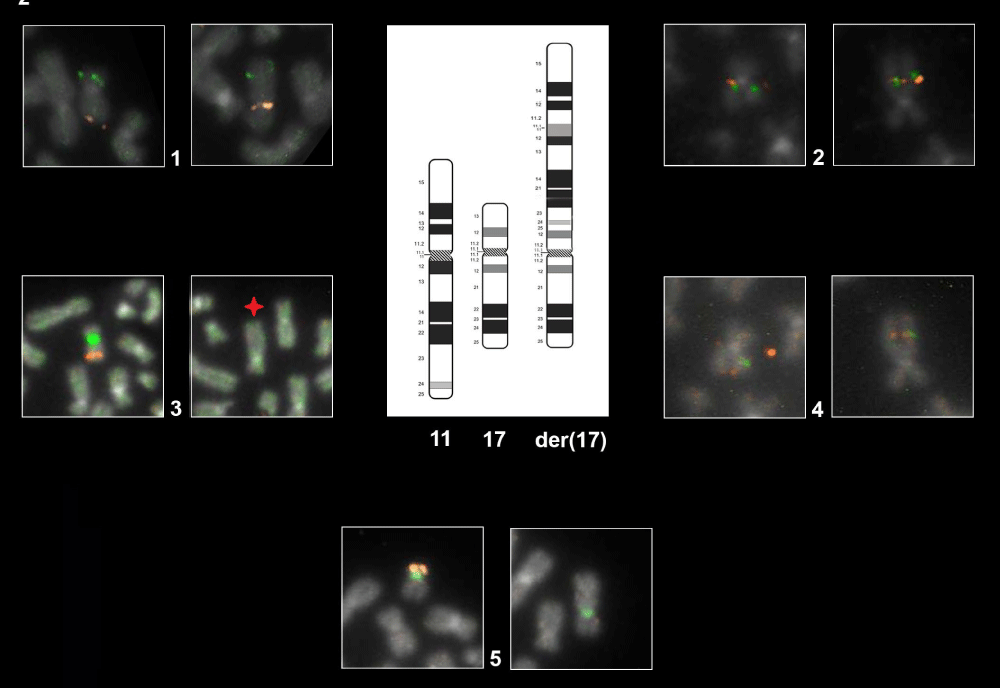
An uncommon translocation t(11;17)(q24;p13) and a concurrent paracentric inversion inv(11)(q13q23) coexisted in the same derivative chromosome as shown in Figures 2-3.
Figure 2: Representative FISH Panel probes. The first eight cells each with two boxes marked with the number 1-2-3-4 show the normal ch.11 (left box) and the der(17) (right box) hybridized with different probes as follow: Box 1: WT1/FLI1 by combination probe set. FISH result highlight the der(17) orientation: green signal (WT1) map on the short arm of chromosome 11 and orange signal (FLI1) map on the long arm of chromosome 11. Box 2: CCND1 Dual Color Break Apart Probe Rearrangement. Der(17) harboring the CCND1 gene in closed proximity to 17p compared with normal chromosome. Box 3 LSI ATM/CEP11. As shown on normal ch.11 (left box) the orange fluorochrome hybridizes ATM gene and the green fluorochrome direct labeled the CEP11. The star on der(17) (right box) show the interstitial deletion affecting the gene ATM and the loss of CEP11. Box 4: MLL Dual Color Break Apart rearrangement. Der(17) harboring the MLL gene in closed proximity to 11p. The last two boxes, marked with the number 5, shows the normal ch.17 (left box) and der(17) in the (right box) hybridized with TP53 (orange)/CEP17(green). As illustrated der(17) keeps intact the CEP17.
Figure 3: Idiograms of G-banding patterns. Representation of the origins of derivative chromosome 17 that arise after structural rearrangements: deletion and paracentric inversion.
To better define the cytogenetic aberration that was difficult to resolve on morphological grounds, FISH analysis was carried out on metaphase plates and interphase nuclei. Five types of Locus Specific Identifier (LSI) probes available in our laboratory were selected in order to mark different regions on the chromosome of interest der(17) as shown in Figure 2. The FISH panel probes used are list as follow: 1) Vysis LSI TP53 SpectrumOrange/CEP17SpectrumGreen Probes: designed for the detection of TP53 gene deletions and its centromere 17 as shown in Figure 2 panel 5; 2) Vysis LSI ATM SpectrumOrange/CEP11SpectrumGreenProbes; the probe was intended to detect the loss of the 11q22 region as shown in Figure 2 panel 3; 3) ZytoLight SPEC CCND1 break-apart probe mapping on 11q13.3 was selected to detect the chromosomal inversion as shown in Figure 2 panel 2; 4) LSI MLL Dual Color Break Apart (Vysis, Abbott) mapping to chromosome 11q23 useful for detecting the chromosomal inversion as shown in Figure 2 panels 4,5) LSI WT1/FLI1 custom FISH combination probes: WT1 has been designed as a one-color probe (green) and mapping to chromosome 11p13; FLI1 probe was labeled with SpectrumOrange and mapping to chromosome 11q24.3 as shown in Figure 2 panel 1.
Therefore, by using each single probe, it was possible to highlight uncommon complex chromosome rearrangements involving more than two chromosomes breaks.
As shown in the Figure 1, the karyotype contained one normal chromosome 17, one normal chromosome 11 and the rearranged chromosome der(17). Derivative(17) was a structurally rearranged chromosome generated from the unbalanced translocation of the whole chromosome 11 to the chromosome 17 that kept intact its centromere. The centromere has been clearly confirmed hybridizing with SPEC TP53/CEN 17 Dual Color Probe on metaphase as shown in Figure 2 panel 5. In the translocation t(11;17)(q24;p13) the long arm 11q “sticked” together on 17p leading to a deletion of the short arm 17p13 (TP53 locus). The orientation of the short and long arms of chromosome 11, following the translocation on chromosome 17, were established by LSI WTI1 and FLI1 FISH probes as shown in Figure 2 panel 1.
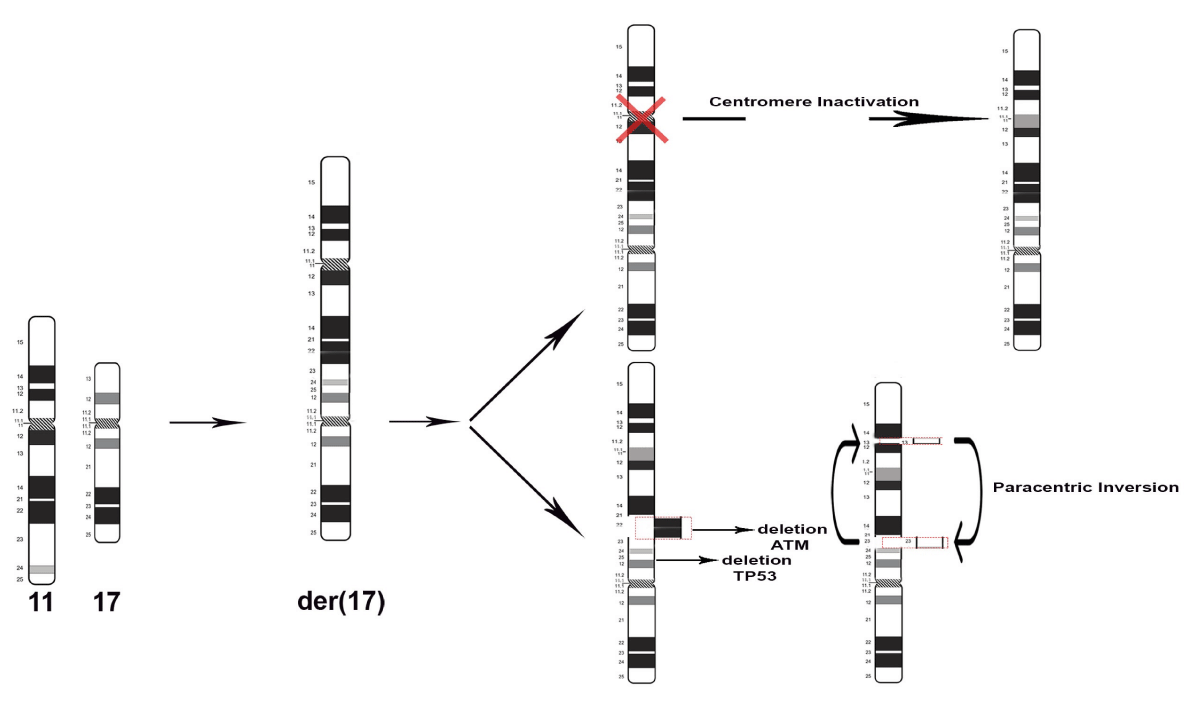
Furthermore, other aberrations have occurred within der(17), like the ATM deletion (11q22) and the paracentric inversion in which breakage and reunion have occurred at bands 11q13 and 11q23 are highlighted in the Figure 3. Overlapping the hybridization signals pattern of MLL and CCND1 on rearranged chromosomes der(17) we observed that CCND1 segment was reversed in relation to MLL. Therefore the CCND1 region was in closed proximity to 17p and viceversa.
To our knowledge, this structural rearrangement was never reported before and it bears high clinical relevance as it leads to the loss of two important genes for CLL prognosis: TP53 gene, which is often involved in the regulation of the cell cycle and apoptosis, and ATM gene, which is involved in the DNA repair pathway. In December 2020, due to Binet stage C CLL a new line of therapy with Venetoclax – Rituximab was started, and, at the last follow-up the patient is in partial response with reduced adenopathies and the improvement of blood cell count.
Genetic instability or changes in the chromosome number or structure are important factors in oncogenesis [4]. Structural alterations are known to participate in the early steps of tumorigenesis and in cancer heterogeneity, but the mitotic errors that lead to structural chromosome alterations still remain elusive [5]. Types of cytogenetic rearrangements that are found in tumors like translocations or inversions affect genes at a limited distance from the point of breakage and can result in the chromosomal deregulation of genes or formation of chimeric oncogenes [4]. Therefore, cytogenetic data indicated that the chromosomal changes in a tumor could be used for tumor classification, diagnosis and prognosis. In this report we described a new chromosome rearrangement in CLL patients der(17)t(11;17)(q24;p13)inv(11)(q13;q23) that might represent a novel feature. Our pseudocentric chromosome 17 involves one break at the peri-centromeric region (17p) leading to loss of genetic material (TP53) and through chromosomal fusion with 11q arm generating dicentrics (Figure 2). As described in Barra and Fachinetti [5] dicentric chromosomes are generally unstable and one of the two centromeres becomes inactive causing loss of centromeric components. Accordingly, the newly generated sub-metacentric derivative chromosome preserve the centromere 17 but loss the centromere of chromosome 11. A hypothetical explanation of our cytogenetic event has been described by Khanna and Stephen [6]. When a double strand (DBS) occurs, the chromosome becomes unstable and needs to be repaired. This problem may be solved by joining the damaged chromosomes, but erroneous rejoining of broken DNA may occur, resulting in the deletion or translocations as well as inversion. It is known that chromosomal aberrations may provide some advantageous phenotypic changes in the tumor cells, enabling them to evade apoptosis by retaining their proliferation potential. Indeed, CCND1 is a partner of recurrent fusions in several hematological malignancies and seems to cooperate with ATM gene as describe in Schaffner, et al. [7]. ATM inactivation in tumor cells may act synergistically with cyclin D1 to override cell-cycle checkpoint controls and promote the accumulation of additional genomic aberration during tumorigenic development. Both ATM and P53 genes are relevant for CLL prognosis, as their mutations or deletions confer chemotherapy resistance to CLL cells [1,2]. Since our patient presented deletion of both genes - at more sensitive analyses than karyotype - before starting first-line therapy, it is impossible to differentiate if the derivative chromosome we described was already present in a minor sub clone at the beginning of the disease history and then expanded upon therapy selective pressure, or if it was generated directly at disease relapse, possibly favored by the dual deletion of chromosome 11 and 17 themselves. In any case, the derivative chromosome we described is highly relevant since it leads to the simultaneous and combined deletion of two highly relevant genes in CLL pathogenesis. In conclusion, cytogenetic techniques are valuable tools to diagnose cancer and to direct cancer treatment. It is our belief that the interface between classic cytogenetics and molecular cytogenetics must be exploited to generate the data to improve our knowledge of cancer biology.
Statement of ethics
We wish to confirm that all investigations were conducted according to the principles expressed in the Declaration of Helsinki.
Consent informed
Written informed consent was obtained from the patient for publication of this case report
Author contribution
Valentina Monti conceived and wrote the manuscript. Fabio Serpenti, Lucia Farina, Maria Adele Testi provided the patient data, clinical information and revised the paper. Giancarlo Pruneri carried out the histopathological diagnosis. Maria Luisa Moiraghi performed the technical tasks in the cytogenetic Laboratory. All the authors read the last version of the manuscript.
- Buccheri V, Barreto WG, Fogliatto LM, Capra M, Marchiani M. Prognostic and therapeutic stratification in CLL: focus on 17p deletion and p53 mutation. Ann Hematol. 2018; 97: 2269-2278. PubMed: https://pubmed.ncbi.nlm.nih.gov/30315344/
- Stankovic T, Skowronska A. The role of ATM mutations and 11q deletions in disease progression in chronic lymphocytic leukemia. Leuk Lymphoma. 2014; 55: 1227-1239. PubMed: https://pubmed.ncbi.nlm.nih.gov/23906020/
- Stilgenbauer S, Lichter P, Dohner H. Genetic features of B cell chronic Lympocytic leukemia. Rev Clin Exp Hematol. 2000; 4: 48-72. PubMed: https://pubmed.ncbi.nlm.nih.gov/11486330/
- Oliveira-Junior RJ, Filho LRG, Bastos LM, Alves DD, Vaz dos Santis S. Contribution of cytogenetics to cancer research. 2014; 30: 245-259.
- Barra V, Fachinetti D. The dark side of centromeres: types, causes and consequences of structural abnormalities implicating centromeric DNA. Nat Commun. 2018; 9: 4340. PubMed: https://pubmed.ncbi.nlm.nih.gov/30337534/
- Khanna KK, Stephen PJ. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001; 27: 247-254. PubMed: https://pubmed.ncbi.nlm.nih.gov/11242102/
- Schaffner C, Idler I, Stilgenbauer S, Dohner H. Lichter P. Mantle cell lymphoma is characterized by inactivation of the ATM gene. Proc Natl Acad Sci USA. 2000; 97: 2773-2778. PubMed: https://pubmed.ncbi.nlm.nih.gov/10706620/
- Shaffer LG, McGowan-Jordan J, Schmid M. An International System for Human Cytogenetic Nomenclature (ISCN). Basel. 2013.